Clustering with a Key-Value Store
Let’s say you have a dataset you’d like to cluster. Let’s say you don’t want to write more than 5 lines of code. Let’s say that your only tool is a key-value store. (Why might you be in this position? Perhaps your dataset is really really (really) big and only simple things will scale. Maybe it’s in fact INFINITE and you’re clustering a stream. Maybe MapReduce is just a really big hammer. 🔨👷 Why you’d only want to write 5 lines of code is left as an exercise to the reader.)
At any rate, you would like to make clusters out of your data, but you only get to look at each item once in isolation. After looking at it you have to decide what cluster it should go to, at that moment, without looking at any other information, or any other items in your dataset. You only get one shot, do not throw it away! How can we accomplish this?
Ideally we want a magic function, where
if and only if
and
should be in the same cluster. We don’t have such a function! (sorry) But we do have something pretty close.
Let’s say you have a function that computes a hash of your item, and your hash has the following property,
for some similarity measure
.
is called a “locality sensitive hash” for
. This is pretty close to we want! Things that are similar to each other will have a high probability of sharing a key, and things that are dissimilar to each other have a low probability of sharing a key. Later we’ll talk about how to make this behave a bit more like our magic function, but for now, lets talk about how to build this one.
// C++ code implementing each algorithm will be in a block like this one // at the end of each section.
Building a Simple H(X): MinHash
Suppose you have two sets, and
, and you would like to know how similar they are. First you might ask, how big is their intersection?
That’s nice, but isn’t comparable across different sizes of sets, so let’s normalize it by the union of the two sizes.
This is called the Jaccard Index, and is a common measure of set similarity. It has the nice property of being 0 when the sets are disjoint, and 1 when they are identical.
Suppose you have a uniform pseudo-random hash function from elements in your set to the range
. For simplicity, assume that the output of
is unique for each input. I’ll use
to denote the set of hashes produced by applying
to each element of
, i.e.
.
Consider . When you insert and delete elements from
, how often does
change?
If you delete from
then
will only change if
. Since any element has an equal chance of having the minimum hash value, the probability of this is
.
If you insert into
then
will only change if
. Again, since any element has an equal chance of having the minimum hash value, the probability of this is
.
For our purposes, this means that is useful as a stable description of
.
What is the probability that ?
If an element produces the minimum hash in both sets on their own, it also produces the minimum hash in their union.
if and only if
. Let
be the member of
that produces the minimum hash value. The probability that
and
share the minimum hash is equivalent to the probability that
is in both
and
. Since any element of
has an equal chance of having the minimum hash value, this becomes
Look familiar? Presto, we now have a Locality Sensitive Hash for the Jaccard Index.
unsigned long long int MinHash(T X) {
unsigned long long int min_hash = ULLONG_MAX;
for (auto x : X) {
min_hash = min(min_hash, Hash(x));
}
return min_hash;
}
Tuning Precision and Recall with Combinatorics
Now that we have a locality sensitive hash, we can use combinatorics to build something that looks a bit more like our magic function. We can concatenate (or sum) hashes to perform an “AND” operation. Let , i.e. concatenating
independent hashes. The probability that
is then
.
We can then output multiple hashes to perform an “OR” operation. If we output independent hashes, then the probability that at least one of those hashes is the same for two items is
.
Using these two tools, we can apply a “sigmoid” function to our similarities, outputting independent copies of
concatenated hashes, the probability that two items will share at least one key is
. (You can think of this as the probability that two items will “meet each other” at least once in the course of your computation.)
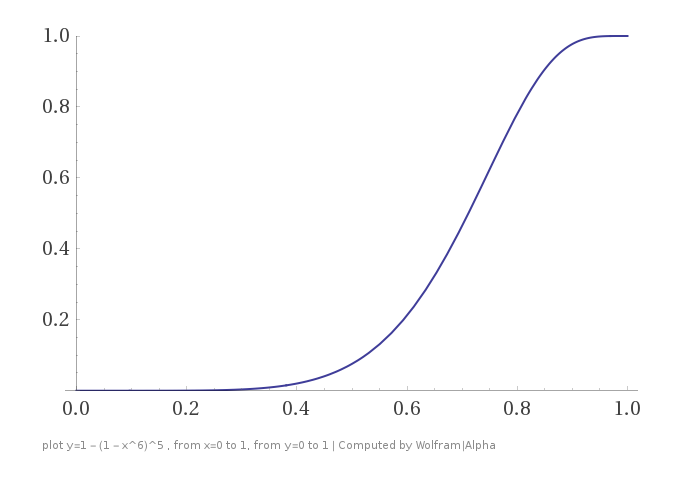
Now we can be pretty sure that things that are similar to each other will share at least one key, and things that aren’t won’t. We can increase the sharpness of the sigmoid as much as we want by spending more storage and CPU to increase A and O.
Great! Clustering with a key-value store! Now let’s talk about ways to improve .
template<T>
unsigned long long int MinHash(T X, int s) {
unsigned long long int min_hash = ULLONG_MAX;
for (auto x : X) {
// Note that the hash function must now accept a seed.
min_hash = min(min_hash, Hash(x, s));
}
return min_hash;
}
template<T>
void EmitWithKeys(T X, int ands, int ors) {
for (int o = 0; o < ors; o++) {
unsigned long long int key = 0;
for (int a = 0; a < ands; a++) {
// we assume that a large int is enough keyspace that we
// can get away with adding instead of concatenating.
key += MinHash(X, a + o * ands);
}
Emit(key, X);
}
}
Integer Weights
This algorithm works on a set, but the things we’d like to cluster usually aren’t sets. For instance, terms in a document (even long n-grams) can occur multiple times and ideally we don’t just want to just discard counts of how often they occur. How can we fix this?
Easy! Just hash everything multiple times. If an element occurs times in your set, hash it
independent times, insert each of them, and then take the minimum of this expanded set as your hash just like before.
We expand our hash function to accept both an item and an integer as its argument, . And if item
occurs
times, then insert
. Now think about what this means for the intersection and the union of these sets. If the count of
in object
is
, and the count of
in object
is
, then the intersection of the two sets of hashes has
items, and the union has
items. You can imagine stacking these hashes on top of each other to form a histogram.
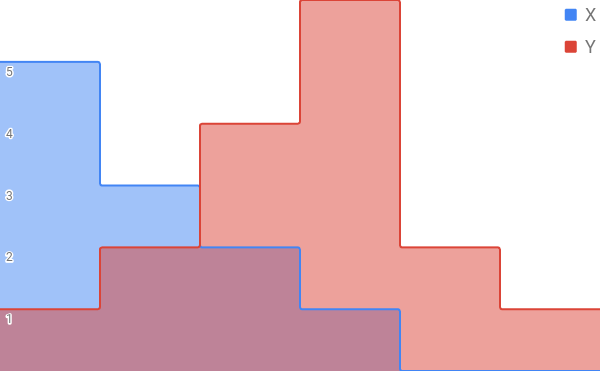
And then when you perform the intersection and union operations, these translate into performing a min and max across the values of each item.
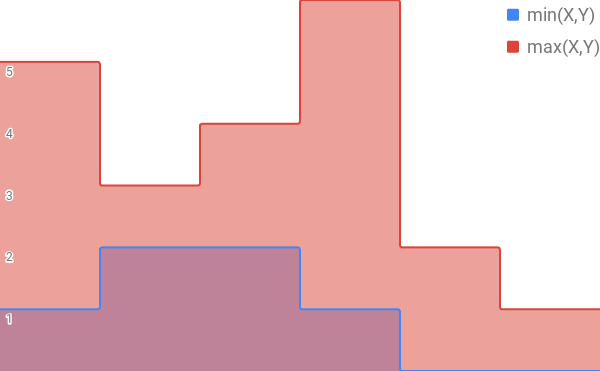
So now that we’re working with vectors instead of sets, if you interpret the Jaccard Index on this expanded set in terms of a weighted vector, it turns into the “Weighted Jaccard Index.”
template<T>
unsigned long long int IntegerWeightMinHash(map<T, int> X, int s) {
unsigned long long int min_hash = ULLONG_MAX;
for (auto x : X) {
for (int i = 0; i < x.second; ++i) {
min_hash = min(min_hash, Hash(x.first, i));
}
}
return min_hash;
}
If you’d like to have as your match probability but with real weights instead of integer weights, there are two good algorithms to do so, one for dense data, and the other for sparse data (like the original MinHash). They are complicated enough that I’m not going to talk about them more here, but not so complicated that you’ll have trouble implementing them from the algorithm definitions in the papers.
Real Weights and Probability Distributions
Instead of going deep into the algorithms for let’s do something simpler. We’ll go back to the first MinHash technique we discussed, and start tweaking it. We started with uniformly distributed hashes, but now we want to bias them in some way. We want items with higher weight to tend to have smaller hashes, so that they’re more likely to be the minimum. Ideally we’d like the probability that they are the minimum to be exactly proportional to their weight. It turns out that this is really easy to do. If
is uniform between 0 and 1, and
is the weight of
then we transform each hash as follows.
Then we take the item with the minimum hash as our representative just like we did before.
The probability that the hashes collide is the “probability jaccard index” which generalizes the Jaccard index to probability distributions. (For a derivation, see our paper on minhashing probability distributions.)
This is a strange looking formula, what is it? And why is it the Jaccard Index of probability distributions? We’ll get to that in the next section. First, let’s figure out why this particular transformation works. It has to do with the beautiful things exponential random variables do when you sort them.
A variable is exponentially distributed if
. It has one parameter,
called its “rate.”
The distribution of the minimum of two random variables is easy to derive from their distributions. If
and
are exponential with rates
then this simplifies to
, another exponential! In addition, the probability that
is
. Proof:
This gives us a new way of sampling from a distribution. If I have a probability distribution with parameters , I can generate a set of independent exponentials with rates
, and then the index with the minimum exponential is a sample from the original distribution.
So, back to minhashing. Instead of generating exponential random variables, now we want to generate “exponentially distributed hashes.” To do that, all you have to do is take a uniformly distributed hash from 0 to 1, invert the exponential CDF, and apply it.
Then if we take the minimum of all these exponentially distributed hashes, we have a sample from the distribution that is stable under small changes, just like the original MinHash!
template<T>
T ProbabilityMinHash(map<T, float> X, int s) {
pair<double, T> min_hash;
min_hash.first = std::numeric_limits::infinity;
double maximum_hash = ULLONG_MAX;
for (auto x : X) {
double zero_one_hash = Hash(x.first) / maximum_hash;
pair<double, T> exponential_hash(-log(zero_one_hash) / x.second, x.first);
min_hash = min(exponential_hash, min_hash);
}
return min_hash.second;
}
Understanding the Probability Jaccard Index
How do you compute the “union” of two probability distributions? How do you compute an “intersection?” One proposal might be to use the and
as we did for the weighted Jaccard index above. This worked for “weighted sets” where the mapping from set to vector is to give every element a weight of either 0 or 1. What happens if you convert these sets to probability distributions instead? In that case we need it to sum to 1, so rather than giving each element a 0 or a 1, we’ll give each element either a 0 or a
. When we do that, the weighted Jaccard index no longer generalizes the Jaccard index, in fact, it’s almost always less. If
and
are sets, with
, and
and
the corresponding uniform probability distributions, then
. Instead, for
,

How can we fix this? We’d like to normalize the distributions in some way so that this no longer happens. Our goal is to make it so that uniform distributions have the same likelihood of colliding as when we applied MinHash to the set that they’re distributed over. Let’s generalize to allow the distributions to be rescaled before we compute each term.
Now, since is too low, let’s choose the vector
to maximize the collision probability we get out. If
, increasing
raises only the denominator. If
, increasing
raises the numerator proportionally more than the denominator. So the optimal
sets
.
 You can derive another representation using the algorithm definition. A set of exponential random variables, when normalized to sum to 1, is a uniformly distributed sample from the unit-simplex. (A unit-simplex is an equilateral triangle in n-dimensions.) In addition, because a unit-simplex is just the set of non-negative vectors that sum to 1, every point on the unit simplex is a probability distribution.
You can derive another representation using the algorithm definition. A set of exponential random variables, when normalized to sum to 1, is a uniformly distributed sample from the unit-simplex. (A unit-simplex is an equilateral triangle in n-dimensions.) In addition, because a unit-simplex is just the set of non-negative vectors that sum to 1, every point on the unit simplex is a probability distribution.
From this, we can represent geometrically.
Take the unit simplex and mark the point on the simplex that corresponds to the distribution you would like to hash. Connect that point to each corner of the simplex with an edge. These edges you’ve drawn split the simplex into smaller simplexes, where each one has area proportional to the weight of one of the elements of your distribution.
With the distribution represented this way, you can imagine sampling from it by throwing darts at the simplex. The id of whichever region the dart lands in is a sample from your distribution. The probability MinHash does exactly this, except that the dart remains in the same spot for each distribution you sample from, and the boundaries of the regions shift around it.
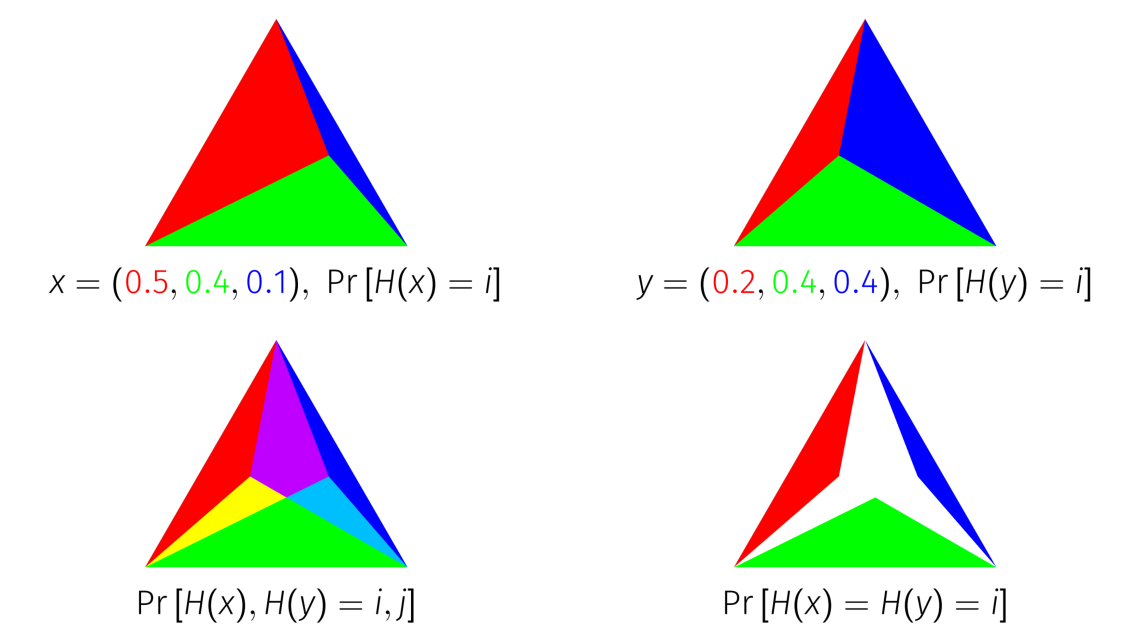
So now, we’re able to visualize on this simplex. When you overlay two distributions on top of each other, and intersect the simplexes of the same color, the area that remains is equal to
.
This representation also lets us demonstrate geometrically the most interesting fact about and the strongest reason to call it the Jaccard Index of Probability Distributions.
Theorem:
is maximal.
For any sampling method, if
, then for some
where
and
, either
or
.
In other words, no method of sampling from discrete distributions can have collision probabilities that are greater than everywhere. If you try to get higher recall on one pair than this algorithm provides, you have to give up recall on at least one “good” pair, a pair that is more similar than the pair you are trying to improve.
This theorem is true of and
but not
! The analogous
for the original MinHash is
. The original MinHash, and the probability MinHash both make
collide as much as possible.
The proof of this in the general case is complicated, but the simplex representation allows us to prove it purely visually on three element distributions.
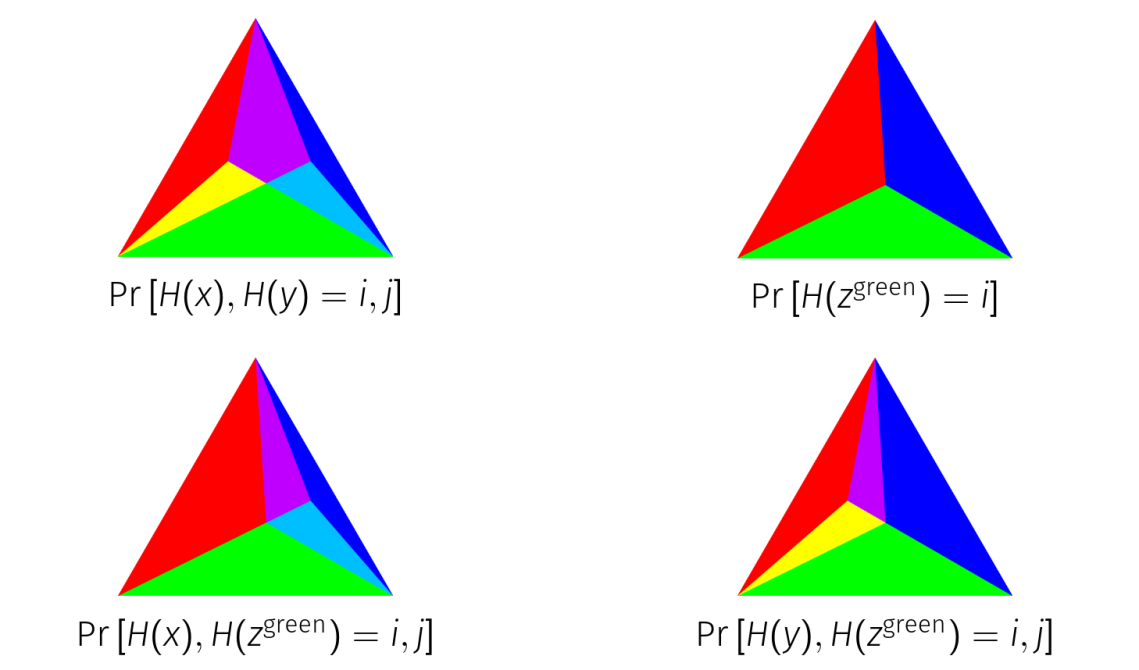
We form a new distribution out of the intersection of the green simplexes. This makes a distribution that is in-between and
but collides with both of them as much as possible on every possible outcome. For
and
to collide more on the green element, they have to give up collisions with
on either red or blue, and can’t gain any more on green to compensate.
For a lot more about this see our Probability Minhash paper.
